Setting up the Phenix GUI to work with SLURM scheduler and rosetta
Phenix and rosetta are two software packages in rapid development that can be used for MX structure solution, autobuilding and refinement of protein crystal structures. Rosetta started as a computational modeling and protein structure analysis software and Phenix integrated rosetta and are now integrating alphaFold and Cryo-EM applications making Phenix a truly integrative structural biology software package.
First check what Phenix versions are available in PReSTO by:
module avail Phenix
where the default module that loads from the PReSTO menu is labeled with a (D). You may also load old/later Phenix versions by “module load …” command
The path to the rosetta software must be set in Phenix preferences - Wizards
Figure 1. Phenix preferences - Wizards. Add rosetta path to Phenix GUI.
OBS OBS Use OLD path to rosetta as in Phenix 1.19.2-4158
Phenix developers recommend using Phenix 1.19.2-4158 with rosetta compared to using 1.20.1-4487. We can confirm this by successfully run a rosetta.refine test job using Phenix/1.19.2-4158-Rosetta-3.10-8-PReSTO that were indeed failing in Phenix/1.20.1-4487-Rosetta-3.10-5-PReSTO. The “path to Rosetta” setting in Phenix GUI should be: /software/presto/software/Phenix/1.19.2-4158-foss-2019b-Rosetta-3.10-8/rosetta-3.10
Using the PReSTO menu, the phenix GUI is launched at the login node. Small single processor phenix jobs can be run at the login node and parallel jobs can be send to the compute nodes in two ways:
- Using the inherent slurm scheduler in Phenix
- Using the “save parameter file” option followed by a standard sbatch script
Option 1 Using the inherent slurm scheduler in Phenix is better because the phenix logfiles and resulting pdb and mtz files is readily visible in the phenix GUI, while if using the “save parameter file” option the phenix-GUI cannot track the output of the standard sbatch script.
Option 1. Using the inherent slurm scheduler in Phenix (preferred)
The Phenix GUI slurm parameters are set under Preferences - Processing and need to be changed frequently since running phenix.refine is less than an hour in most cases while phenix.mr_rosetta could take more 24 hours when using all cores of a compute node.
Figure 2. The inherent phenix GUI slurm scheduler. The Queue submit command need to be frequently edit to match the time it take for a job to finish. If always using 48 hours, a typical 1-hour phenix.refine job might wait for a long time in the queue before being launched at a compute node
The Queue submit command is a typical sbatch script header written in a single line like for short phenix.refine jobs
sbatch –nodes=1 –exclusive -t 1:00:00 -A naiss2023-22-811 –mail-user=name.surname@lu.se –mail-type=ALL
and for much longer phenix.mr_rosetta jobs
sbatch –nodes=1 –exclusive -t 48:00:00 -A naiss2023-22-811 –mail-user=name.surname@lu.se –mail-type=ALL
After editing suitable settings in Preferences - Processes the most general way to submit a job directly from the is via Actions - Queueing system - Submit job as indicated in figure 2.
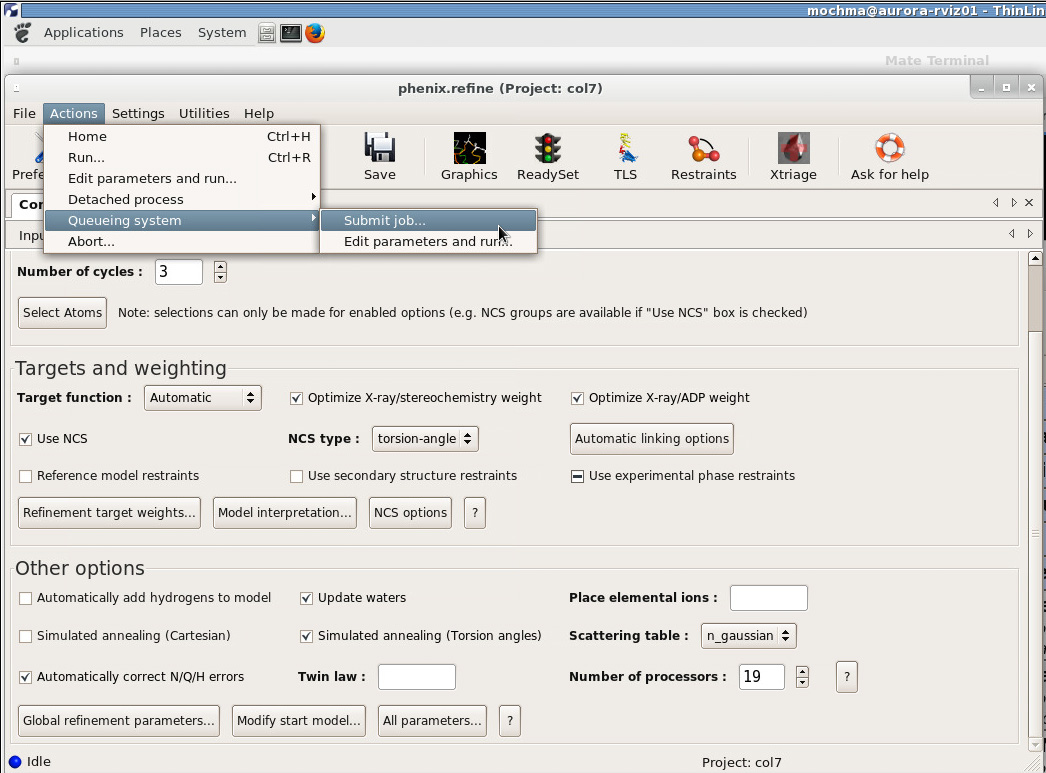
Figure 3. Sending a parallel job to the queue using the slurm parameters specified under Preferences - Processes.
Importantly the Number of processors should match the number of cores available at a single compute node minus one core, so use 47 for Cosmos and 31 for Tetralith. Multi-node usage of phenix.mr_rosetta is possible, however not recommended at present the savings in wall-clock time is very limited and therefore a waste of compute time. When running Phenix-GUI at BioMAX, the login node that has 40 cores and compute nodes have 20 cores at offline-fe1 (and 24 cores at clu0-fe-1). This mean you must adapt Number of processes from 39 to 19 at offline-fe1 (or 23 at clu0-fe-1)
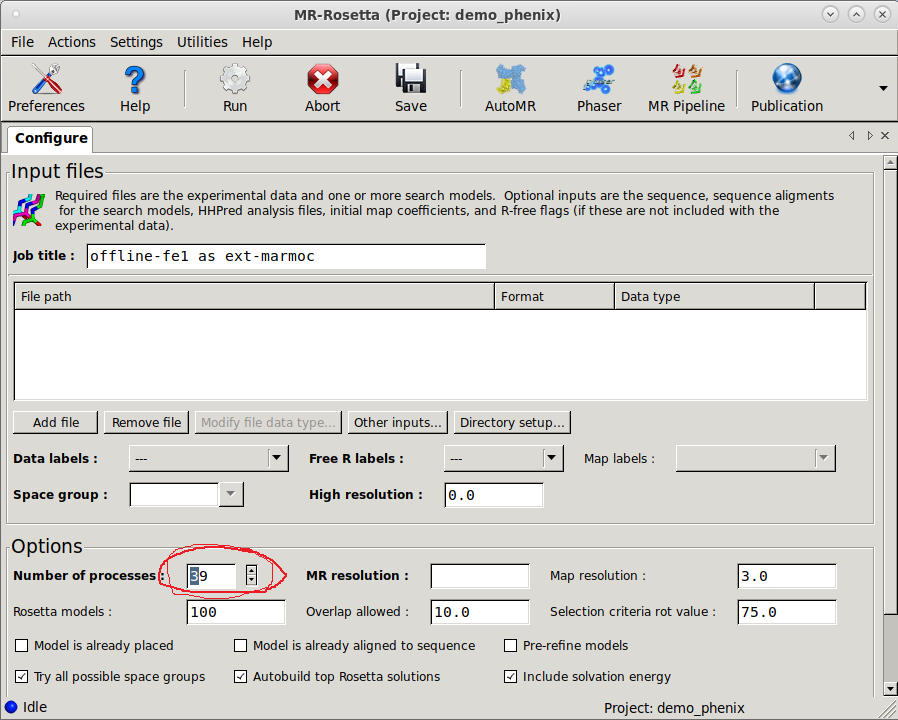
Figure 4. Change 39 to 19 at offline-fe1(offline cluster), 23 at clu0-fe-1(online cluster) and 47 at LUNARC Cosmos
Option 2. Using the “save parameter file” option followed by a standard sbatch script
After editing the phenix wizard save the parameter file as indicated in figure 3.
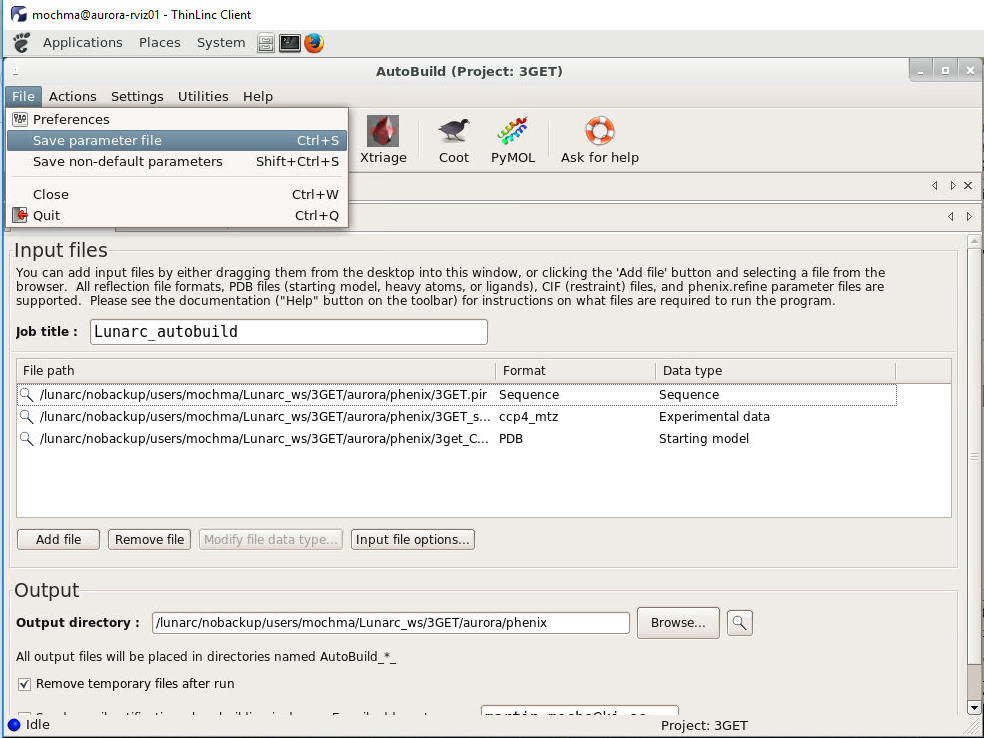
Figure 5. How to save parameter file from phenix GUI.
After saving the parameter file, submit a sbatch script
Phenix autobuild sbatch script
#!/bin/sh
#SBATCH -t 2:00:00
#SBATCH -N 1 --exclusive
#SBATCH -A naiss2023-22-811
#SBATCH --mail-type=ALL
#SBATCH --mail-user=name.surname@lu.se
module load Phenix
phenix.autobuild autobuild_1.eff
Phenix MR rosetta sbatch script
#!/bin/sh -eu
#SBATCH -t 18:00:00
#SBATCH --nodes=1 --exclusive
#SBATCH -A naiss2023-22-811
#SBATCH --mail-type=ALL
#SBATCH --mail-user=name.surname@lu.se
module load Phenix/1.17.1-3660-Rosetta-3.10-5-PReSTO
phenix.mr_rosetta mr_rosetta_5.eff
Phenix mail settings
Phenix-GUI e-mail settings at NSC Tetralith and LUNARC Cosmos. Phenix-GUI cannot send emails when run at MAXIV cluster.
Figure 6. Phenix GUI email settings for Karolinska Institutet user.
 User Area
User Area

